How to’s¶
Loading run data¶
Reading the bunch pattern¶
Extracting peaks from digitizers¶
Determining the FEL or OL beam size and the fluence¶
Finding time overlap by transient reflectivity¶
Transient reflectivity of the optical laser measured on a large bandgap material pumped by the FEL is often used at SCS to find the time overlap between the two beams. The example notebook
shows how to analyze such data, including correcting the delay by the bunch arrival monitor (BAM).
DSSC¶
DSSC data binning¶
In scattering experiment one typically wants to bin DSSC image data versus time delay between pump and probe or versus photon energy. After this first data reduction steps, one can do azimuthal integration on a much smaller amount of data.
The DSSC data binning procedure is based on the notebook Dask DSSC module binning. It performs DSSC data binning against a coordinate specified by xaxis which can be nrj for the photon energy, delay in which case the delay stage position will be converted in picoseconds and corrected but the BAM, or another slow data channel. Specific pulse pattern can be defined, such as:
['pumped', 'unpumped']
which will be repeated. XGM data will also be binned similarly to the DSSC data.
Since this data reduction step can be quite time consuming for large datasets,
it is recommended to launch the notebook via a SLURM script. The script can be
downloaded from scripts/bin_dssc_module_job.sh and reads as:
1#!/bin/bash
2#SBATCH -N 1
3#SBATCH --partition=exfel
4#SBATCH --time=12:00:00
5#SBATCH --mail-type=END,FAIL
6#SBATCH --output=logs/%j-%x.out
7
8while getopts ":p:d:r:k:m:x:b:" option
9do
10 case $option in
11 p) PROPOSAL="$OPTARG";;
12 d) DARK="$OPTARG";;
13 r) RUN="$OPTARG";;
14 k) KERNEL="$OPTARG";;
15 m) MODULE_GROUP="$OPTARG";;
16 x) XAXIS="$OPTARG";;
17 b) BINWIDTH="$OPTARG";;
18 \?) echo "Unknown option"
19 exit 1;;
20 :) echo "Missing option for input flag"
21 exit 1;;
22 esac
23done
24
25# Load xfel environment
26source /etc/profile.d/modules.sh
27module load exfel exfel-python
28
29echo processing run $RUN
30PDIR=$(findxfel $PROPOSAL)
31PPROPOSAL="p$(printf '%06d' $PROPOSAL)"
32RDIR="$PDIR/usr/processed_runs/r$(printf '%04d' $RUN)"
33mkdir $RDIR
34
35NB='Dask DSSC module binning.ipynb'
36
37# kernel list can be seen from 'jupyter kernelspec list'
38if [ -z "${KERNEL}" ]; then
39 KERNEL="toolbox_$PPROPOSAL"
40fi
41
42python -c "import papermill as pm; pm.execute_notebook(\
43 '$NB', \
44 '$RDIR/output$MODULE_GROUP.ipynb', \
45 kernel_name='$KERNEL', \
46 parameters=dict(proposalNB=int('$PROPOSAL'), \
47 dark_runNB=int('$DARK'), \
48 runNB=int('$RUN'), \
49 module_group=int('$MODULE_GROUP'), \
50 path='$RDIR/', \
51 xaxis='$XAXIS', \
52 bin_width=float('$BINWIDTH')))"
It is launched with the following:
sbatch ./bin_dssc_module_job.sh -p 2719 -d 180 -r 179 -m 0 -x delay -b 0.1
sbatch ./bin_dssc_module_job.sh -p 2719 -d 180 -r 179 -m 1 -x delay -b 0.1
sbatch ./bin_dssc_module_job.sh -p 2719 -d 180 -r 179 -m 2 -x delay -b 0.1
sbatch ./bin_dssc_module_job.sh -p 2719 -d 180 -r 179 -m 3 -x delay -b 0.1
where 2719 is the proposal number, 180 is the dark run number, 179 is the run nummber and 0, 1, 2 and 3 are the 4 module group, each job processing a set of 4 DSSC module, delay is the bin axis and 0.1 is the bin width.
The result will be 16 *.h5 files, one per module, saved in the folder specified in the script, a copy of which can be found in the scripts folder in the toolbox source. This files can then be loaded and combined with:
import xarray as xr
data = xr.open_mfdataset(path + '/*.h5', parallel=True, join='inner')
DSSC azimuthal integration¶
Azimuthal integration can be performed with pyFAI which can utilize the hexagonal pixel shape information from the DSSC geometry to split the intensity in a pixel in the bins covered by it. An example notebook Azimuthal integration of DSSC with pyFAI.ipynb is available.
A second example notebook DSSC scattering time-delay.ipynb demonstrates how to:
refine the geometry such that the scattering pattern is centered before azimuthal integration
perform azimuthal integration on a time delay dataset with
xr.apply_ufuncfor multiprocessing.plot a two-dimensional map of the scattering change as function of scattering vector and time delay
integrate certain scattering vector range and plot a time trace
DSSC fine timing¶
When DSSC is reused after a period of inactivity or when the DSSC gain setting use a different operation frequency the DSSC fine trigger delay needs to be checked. To analysis runs recorded with different fine delay, one can use the notebook DSSC fine delay with SCS toolbox.ipynb.
DSSC quadrant geometry¶
To check or refined the DSSC geometry or quadrants position, the following notebook can be used DSSC create geometry.ipynb.
Legacy DSSC binning procedure¶
Most of the functions within toolbox_scs.detectors can be accessed directly. This is useful during development, or when working in a non-standardized way, which is often neccessary during data evaluation. For frequent routines there is the possibility to use dssc objects that guarantee consistent data structure, and reduce the amount of recurring code within the notebook.
bin data using toolbox_scs.tbdet -> to be documented.
post processing, data analysis -> to be documented
Photo-Electron Spectrometer (PES)¶
BOZ: Beam-Splitting Off-axis Zone plate analysis¶
The BOZ analysis consists of 4 notebooks and a script. The first notebook BOZ analysis part I.a Correction determination is used to determine all the necessary correction, that is the flat field correction from the zone plate optics and the non-linearity correction from the DSSC gain. The inputs are a dark run and a run with X-rays on three broken or empty membranes. For the latter, an alternative is to use pre-edge data on an actual sample. The result is a JSON file that contains the flat field and non-linearity correction as well as the parameters used for their determination such that this can be reproduced and investigated in case of issues. The determination of the flat field correction is rather quick, few minutes and is the most important correction for the change in XAS computed from the -1st and +1st order. For quick correction of the online preview one can bypass the non-linearity calculation by taking the JSON file as soon as it appears. The determination of the non-linearity correction is a lot longer and can take some 2 to 8 hours depending on the number of pulses in the train. For this reason, the computation can also be done on GPUs in 30min instead. A GPU notebook adapted for CHEM experiment with liquid jet and normalization implement for S K-edge is available at OnlineGPU BOZ analysis part I.a Correction determination S K-egde.
The other option is to use a script
that can be downloaded from scripts/boz_parameters_job.sh and
reads as:
1#!/bin/bash
2#SBATCH -N 1
3#SBATCH --partition=allgpu
4#SBATCH --constraint=V100
5#SBATCH --time=2:00:00
6#SBATCH --mail-type=END,FAIL
7#SBATCH --output=logs/%j-%x.out
8
9ROISTH='1'
10SATLEVEL='500'
11MODULE='15'
12
13while getopts ":p:d:r:k:g:t:s:m:" option
14do
15 case $option in
16 p) PROPOSAL="$OPTARG";;
17 d) DARK="$OPTARG";;
18 r) RUN="$OPTARG";;
19 k) KERNEL="$OPTARG";;
20 g) GAIN="$OPTARG";;
21 t) ROISTH="$OPTARG";;
22 s) SATLEVEL="$OPTARG";;
23 m) MODULE="$OPTARG";;
24 \?) echo "Unknown option"
25 exit 1;;
26 :) echo "Missing option for input flag"
27 exit 1;;
28 esac
29done
30
31# Load xfel environment
32source /etc/profile.d/modules.sh
33module load exfel exfel-python
34
35echo processing run $RUN
36PDIR=$(findxfel $PROPOSAL)
37PPROPOSAL="p$(printf '%06d' $PROPOSAL)"
38RDIR="$PDIR/usr/processed_runs/r$(printf '%04d' $RUN)"
39mkdir $RDIR
40
41NB='BOZ analysis part I.a Correction determination.ipynb'
42
43# kernel list can be seen from 'jupyter kernelspec list'
44if [ -z "${KERNEL}" ]; then
45 KERNEL="toolbox_$PPROPOSAL"
46fi
47
48python -c "import papermill as pm; pm.execute_notebook(\
49 '$NB', \
50 '$RDIR/output.ipynb', \
51 kernel_name='$KERNEL', \
52 parameters=dict(proposal=int('$PROPOSAL'), \
53 darkrun=int('$DARK'), \
54 run=int('$RUN'), \
55 module=int('$MODULE'), \
56 gain=float('$GAIN'), \
57 rois_th=float('$ROISTH'), \
58 sat_level=int('$SATLEVEL')))"
It uses the first notebook and is launched via slurm:
sbatch ./boz_parameters_job.sh -p 2937 -d 615 -r 614 -g 3
where 2937 is the proposal run number, where 615 is the dark run number, 614 is the run on 3 broken membranes and 3 is the DSSC gain in photon per bin. The proposal run number is defined inside the script file.
The second notebook BOZ analysis part I.b Correction validation can be used to check how well the calculated correction still work on a characterization run recorded later, i.e. on 3 broken membrane or empty membranes.
The third notebook BOZ analysis part II.1 Small data then use the JSON correction file to load all needed corrections and process an run, saving the rois extracted DSSC as well as aligning them to photon energy and delay stage in a small data h5 file.
That small data h5 file can then be loaded and the data binned to compute a spectrum or a time resolved XAS scan using the fourth and final notebook BOZ analysis part II.2 Binning
Point detectors¶
Detectors that produce one point per pulse, or 0D detectors, are all handled in a similar way. Such detectors are, for instance, the X-ray Gas Monitor (XGM), the Transmitted Intensity Monitor (TIM), the electron Bunch Arrival Monitor (BAM) or the photo diodes monitoring the PP laser.
HRIXS¶
Viking spectrometer¶
SLURM, sbatch, partition, reservation¶
Scripts launched by sbatch command can employ magic cookie with
#SBATCH to pass options SLURM, such as which partition to run on.
To work, the magic cookie has to be at the beginning of the line.
This means that:
to comment out a magic cookie, adding another “#” before it is sufficient
to comment a line to detail what the option does, it is best practice to put the comment on the line before
Reserved partition are of the form “upex_003333” where 3333 is the proposal
number. To check what reserved partition are existing, their start and end
date, one can ssh to max-display and use the command sview.
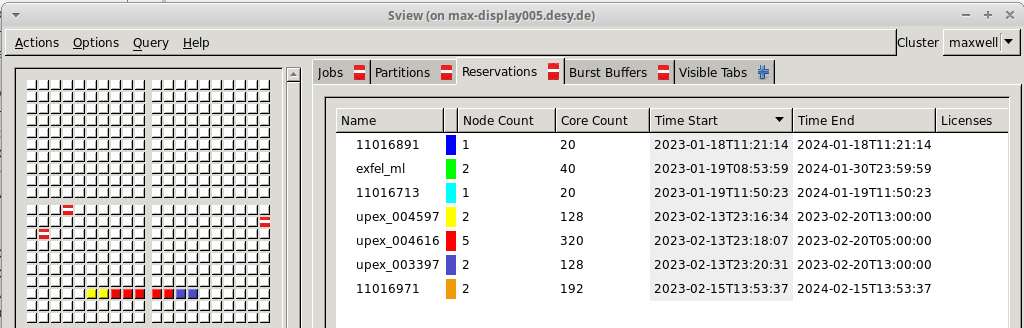
To use a reserved partition with sbatch, one can use the magic cookie
#SBATCH --reservation=upex_003333
instead of the usual
#SBATCH --partition=upex